Publications
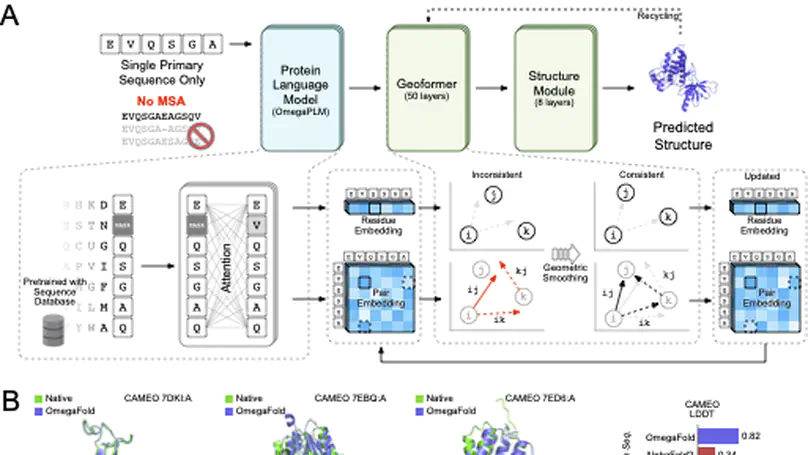
Recent breakthroughs have used deep learning to exploit evolutionary information in multiple sequence alignments (MSAs) to accurately predict protein structures. However, MSAs of homologous proteins are not always available, such as with orphan proteins or fast-evolving proteins like antibodies, and a protein typically folds in a natural setting from its primary amino acid sequence into its three-dimensional structure, suggesting that evolutionary information and MSAs should not be necessary to predict a protein’s folded form. Here, we introduce OmegaFold, the first computational method to successfully predict high-resolution protein structure from a single primary sequence alone. Using a new combination of a protein language model that allows us to make predictions from single sequences and a geometry-inspired transformer model trained on protein structures, OmegaFold outperforms RoseTTAFold and achieves similar prediction accuracy to AlphaFold2 on recently released structures. OmegaFold enables accurate predictions on orphan proteins that do not belong to any functionally characterized protein family and antibodies that tend to have noisy MSAs due to fast evolution. Our study fills a much-encountered gap in structure prediction and brings us a step closer to understanding protein folding in nature.
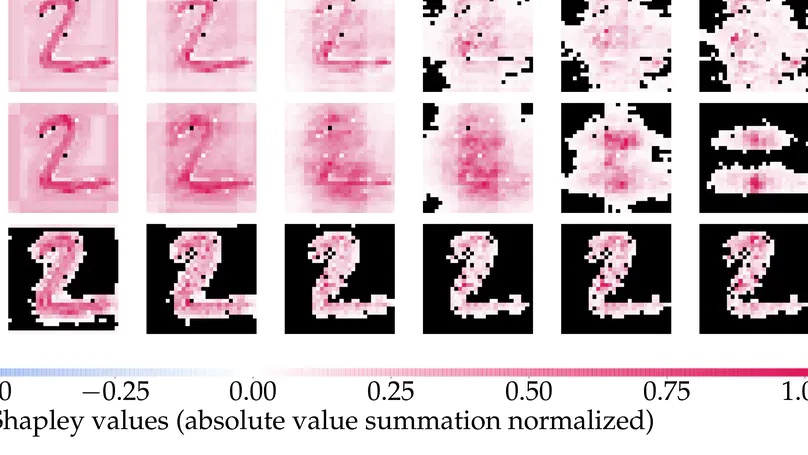
Shapley values have become one of the most popular feature attribution explanation methods. However, most prior work has focused on post-hoc Shapley explanations, which can be computationally demanding due to its exponential time complexity and preclude model regularization based on Shapley explanations during training. Thus, we propose to incorporate Shapley values themselves as latent representations in deep models thereby making Shapley explanations first-class citizens in the modeling paradigm. This intrinsic explanation approach enables layer-wise explanations, explanation regularization of the model during training, and fast explanation computation at test time. We define the Shapley transform that transforms the input into a Shapley representation given a specific function. We operationalize the Shapley transform as a neural network module and construct both shallow and deep networks, called ShapNets, by composing Shapley modules. We prove that our Shallow ShapNets compute the exact Shapley values and our Deep ShapNets maintain the missingness and accuracy properties of Shapley values. We demonstrate on synthetic and real-world datasets that our ShapNets enable layer-wise Shapley explanations, novel Shapley regularizations during training, and fast computation while maintaining reasonable performance. Code is available at https://github.com/inouye-lab/ShapleyExplanationNetworks.